The key to GMOs: XNA
- skhiomura
- Jul 25, 2020
- 7 min read
Genetically modified organisms, more commonly known as GMOs, are fairly common in our daily lives, mainly in the form of genetically modified crops. However, these products have faced pushback from the public for various reasons. One reason opponents against GMOs raise is the potential impact on the environment, such as the unintended creation of pesticide-resistant weeds. One method of avoiding this issue of unwanted genetic pollution is the use of Xeno nucleic acids (XNA). XNAs are “nucleic acid polymers whose backbones are constructed from sugars that are distinct from the natural deoxyribose and ribose sugars found in DNA and RNA” and offer a promising new platform to develop new recombinant organisms without the risk of disrupting the environment. (1) Given that DNA and RNA are fine-tuned for information storage and not necessarily towards being the building blocks of novel nanostructures or the construction of synthetic genetic polymers, it is only natural that the next step would be to create a new platform to pursue these goals. (2)
While similar research was going on before, the term “XNA” was first coined by a paper from Herdewijn and Marliére which accelerated research in this area. (3) In their paper, they defined XNAs as a “type of nucleic acid whose chemical backbone motif would differ from deoxyribose and ribose and whose polymerization would not interfere with DNA and RNA biosynthesis “, formed using nutrients and specific polymerization enzymes so as to prevent them surviving outside controlled environments. (3) This theoretical paper goes further to give four examples of what the new backbone motif could look like, namely anhydrohexitol (HNA), Therose (TNA), Glycerol (GNA), and cyclohexene (CeNA). (3) The paper delves deep into HNA as a promising XNA as HNA polymerase activity was observed for DNA polymerase I in E.coli and for Thermus aquaticus polymerase, where hNTPs (hexitol nucleoside triphosphates) are used to create HNA showing hope that a novel HNA replication system can be created. (3) GNA, TNA have also shown signs of promise, as they have been able to successfully polymerize their sugar-modified nucleoside triphosphate using polymerases as a catalyst on a DNA template(meaning successful GNA/TNA synthesis), while DNA polymerase B and HIV-1 reverse transcriptase showed good CeNA polymerase activity that used DNA as a template, as well as good DNA polymerase activity that used CeNA as a template. (3,4) After this paper was published numerous papers were published regarding the synthesis of XNA. (5,6) While there have been multiple methods of XNA synthesis that have been explored such as solution polymerization, unit-by-unit synthesis and template-directed synthesis, the use of enzymes in synthesizing XNAs has been shown to be a promising new method.(7–10)
In 2012, a paper by Pinheiro et al. explored the synthesis and replication of six XNAs (HNA,TNA, CeNA, locked nucleic acids (LNA), arabinonucleic acids (ANA) and 2’-fluoro-arabinonucleic acids(FANA)) with the use of enzymes(polymerases and reverse transcriptases). (5) The main problem with using polymerases is that the substrate selectivity is very specific making it hard to use them for modified nucleotides such as in the case of XNA synthesis. (5) Therefore it is necessary to engineer polymerases and reverse transcriptases (so as to transcribe the XNA back to DNA) that can accommodate these modified nucleotides. (5) The paper utilized a new method called compartmentalized self-tagging, where polymerases are expressed in compartmentalized bacteria which are exposed to selection primers and then the polymerases bonded to the primers are recovered using paramagnetic beads, that helped to find the right mutations to polymerases and reverse transcriptases that would enable synthesis and reverse transcription of XNA. (5,11) In the case of HNA, the Pol6G12 polymerase was found to display DNA templated HNA polymerase activity, while a mutated reverse transcriptase enzyme known as RT521 was a proficient HNA reverse transcriptase. (5) As for the other XNAs, polymerases PolC7 and PolD4K were efficient in synthesizing CeNA, LNA, ANA, and FANA while the Therminator polymerase was a good synthetase for TNA. (5) RT521 was an efficient reverse transcriptase for TNA, ANA, and FANA, while RT521K (a variant of RT521) was found to enhance CeNA reverse transcriptase activity and enable the reverse transcription of LNA. (5) Out of the six XNAs, TNA has been thoroughly studied, mainly due to how simple the modified backbone (threose) is, making it a very promising medium to store information. (6,12–15)
More recently, a study by Liu et al. in 2018 has looked further into the concept of creating a fully orthogonal system, separate from that of DNA and RNA. (6) In their study, a rather unusual XNA called 3’-2’ phosphonomethyl-threosyl nucleic acid (tPhoNA). (6) tPhoNA was chosen as the subject of the paper due to its orthogonality to DNA and RNA(tying back to the original definition of XNAs by Herdewijn and Marliére) as well as its higher tolerance to degradation by cellular enzyme due to its phosphonate linkage.(3,6) There is great stress on the chemical and biological orthogonality of the XNA, as many XNAs explored in the past have retained “significant affinity for DNA and RNA and substantial efficacy in in vivo transliteration experiments” as well as the fact that “all engineered XNA polymerases so far described also retain substantial DNA polymerase capability”. (6) The study found that tPhoNA could be synthesized using tPhoNA synthetase, which was able to produce a 57-mer in less than 5 minutes using low concentrations of polymerase and manganese. (6) Polymerases RT521 and RT521K, from the study by Pinheiro et al., were found to be efficient reverse transcriptases, although a drop in hybridization between tPhoNA and DNA was admitted when a certain polymerase was absent. (6,16) tPhoNA was also evaluated as a template for DNA synthesis inside of E. coli, where the E. coli colonies that had accepted the tPhoNA as a replication template would survive, where the results showed that E. coli colonies in which took up the tPhoNA had a decrease in prototrophic transformants(an expressed recombinant cells that takes the same nutrients as the original). (6) This is favorable towards the end goal of creating a fully orthogonal system parallel to that of DNA and RNA as this means the XNA is not readily recognized by polymerases designed for DNA.(6) The study shows a shifting goal from the paper from Herdewijn and Marliére in 2009, from creating an orthogonal system that can still communicate with DNAs and RNAs to a fully orthogonal system with minimal interaction to the original nucleic acids and polymerases. (6)
As the amount of research into XNAs has increased dramatically since the paper in 2009, conflicts have arisen inside this research area as well. A paper was published by Chaput and Herdewijn in 2019 addressing two key issues. (1) The first issue was the differing definitions of XNA.(1) In the original paper in 2009, XNA was defined as a “type of nucleic acid whose chemical backbone motif would differ from deoxyribose and ribose and whose polymerization would not interfere with DNA and RNA biosynthesis “. (3) However, in some papers some chemically modified DNAs were listed as XNAs, making the distinction between the two a bit blurred. A good example of this is the paper by Pinheiro et al. above, specifically on how LNA is described. (5) While LNA differs from DNA in its structure, it does not differ enough from its natural counterpart to be considered an “XNA” (compared to DNA, LNA just has an extra oxygen molecule with 2 additional bonds). (1) The second issue is the unclear nomenclature/naming scheme of XNAs. (1) For example, ANA could both stand for arabinose nucleic acid, as well as altritol nucleic acid under the current scheme. (1) The paper proposes a similar system to the well-established carbohydrate nomenclature, making araNA stand for arabinose nucleic acid while altritol nucleic acid would be abbreviated to altNA. (1)
XNAs are a promising orthogonal platform inside of organisms that could be programmed to do tasks without hindering the ability of the organism’s natural enzymes and nucleic acids. Since its popularization in 2009, research in the field has exponentially increased leading to the complete synthesis and reverse transcription of six XNAs in three years and the complete synthesis and reverse transcription of a more orthogonal XNA platform in 9 years.(5,6) While problems do remain, such as the lack of consensus on the definition of XNAs and the lack of clear nomenclature, the topic still gives hope for safer genetically modified organisms and novel medical therapies. (1)
References
1. Chaput JC, Herdewijn P. What Is XNA? Angew Chemie Int Ed [Internet]. 2019 Aug 19 [cited 2020 Jul 19];58(34):11570–2. Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/anie.201905999
2. Pinheiro VB, Holliger P. Towards XNA nanotechnology: New materials from synthetic genetic polymers. Vol. 32, Trends in Biotechnology. Elsevier Ltd; 2014. p. 321–8.
3. Herdewijn P, Marlière P. Toward Safe Genetically Modified Organisms through the Chemical Diversification of Nucleic Acids. Chem Biodivers [Internet]. 2009 Jun 1 [cited 2020 Jul 18];6(6):791–808. Available from: http://doi.wiley.com/10.1002/cbdv.200900083
4. Kempeneers V, Renders M, Froeyen M, Herdewijn P. Investigation of the DNA-dependent cyclohexenyl nucleic acid polymerization and the cyclohexenyl nucleic acid-dependent DNA polymerization. Nucleic Acids Res [Internet]. 2015;Volume 33(12). Available from: https://academic.oup.com/nar/article-abstract/33/12/3828/2400978
5. Pinheiro VB, Taylor AI, Cozens C, Abramov M, Renders M, Zhang S, et al. Synthetic genetic polymers capable of heredity and evolution. Science (80- ) [Internet]. 2012 Apr 20 [cited 2020 Jul 20];336(6079):341–4. Available from: https://science.sciencemag.org/content/336/6079/341
6. Liu C, Cozens C, Jaziri F, Rozenski J, Maréchal A, Dumbre S, et al. Phosphonomethyl Oligonucleotides as Backbone-Modified Artificial Genetic Polymers. J Am Chem Soc [Internet]. 2018 May 30 [cited 2020 Jul 20];140(21):6690–9. Available from: https://pubs.acs.org/sharingguidelines
7. Fairbanks BD, Culver HR, Mavila S, Bowman CN. Towards High-Efficiency Synthesis of Xenonucleic Acids. Vol. 2, Trends in Chemistry. Cell Press; 2020. p. 43–56.
8. Schrum JP, Ricardo A, Krishnamurthy M, Blain JC, Szostak JW. Efficient and rapid template-directed nucleic acid copying using 2′-amino-2′,3′-dideoxyribonucleoside-5′- phosphorimidazolide monomers. J Am Chem Soc [Internet]. 2009 Oct 14 [cited 2020 Jul 19];131(40):14560–70. Available from: https://pubs.acs.org/sharingguidelines
9. Liu ZC, Shin DS, Shokouhimehr M, Lee KN, Yoo BW, Kim YK, et al. Light-directed synthesis of peptide nucleic acids (PNAs) chips. Biosens Bioelectron. 2007 Jun 15;22(12):2891–7.
10. Xi W, Pattanayak S, Wang C, Fairbanks B, Gong T, Wagner J, et al. Clickable Nucleic Acids: Sequence-Controlled Periodic Copolymer/Oligomer Synthesis by Orthogonal Thiol-X Reactions. Angew Chemie Int Ed [Internet]. 2015 Nov 23 [cited 2020 Jul 19];54(48):14462–7. Available from: http://doi.wiley.com/10.1002/anie.201506711
11. Pinheiro VB, Arangundy-Franklin S, Holliger P. Compartmentalized self-tagging for in vitro-directed evolution of XNA polymerases. Curr Protoc Nucleic Acid Chem. 2014;2014:9.9.1-9.9.18.
12. Schoning KU, Scholz P, Guntha S, Wu X, Krishnamurthy R, Eschenmoser A. Chemical etiology of nucleic acid structure: The α-threofuranosyl-(3’→2’) oligonucleotide system. Science (80- ) [Internet]. 2000 Nov 17 [cited 2020 Jul 20];290(5495):1347–51. Available from: https://pubmed.ncbi.nlm.nih.gov/11082060/
13. Yu H, Zhang S, Chaput JC. Darwinian evolution of an alternative genetic system provides support for TNA as an RNA progenitor. Nat Chem [Internet]. 2012 Mar [cited 2020 Jul 20];4(3):183–7. Available from: https://pubmed.ncbi.nlm.nih.gov/22354431/
14. Schöning K-U, Scholz P, Wu X, Guntha S, Delgado G, Krishnamurthy R, et al. The -L-Threofuranosyl-(3′→2′)-oligonucleotide System (‘TNA’): Synthesis and Pairing Properties. Helv Chim Acta [Internet]. 2002 Dec 1 [cited 2020 Jul 20];85(12):4111–53. Available from: http://doi.wiley.com/10.1002/hlca.200290000
15. Ichida JK, Horhota A, Zou K, Mclaughlin LW, Szostak JW. High fidelity TNA synthesis by Therminator polymerase. [cited 2020 Jul 20]; Available from: https://academic.oup.com/nar/article-abstract/33/16/5219/2401165
16. Tsai CH, Chen J, Szostak JW. Enzymatic synthesis of DNA on glycerol nucleic acid templates without stable duplex formation between product and template. Proc Natl Acad Sci U S A [Internet]. 2007 Sep 11 [cited 2020 Jul 23];104(37):14598–603. Available from: www.pnas.org/cgi/content/full/
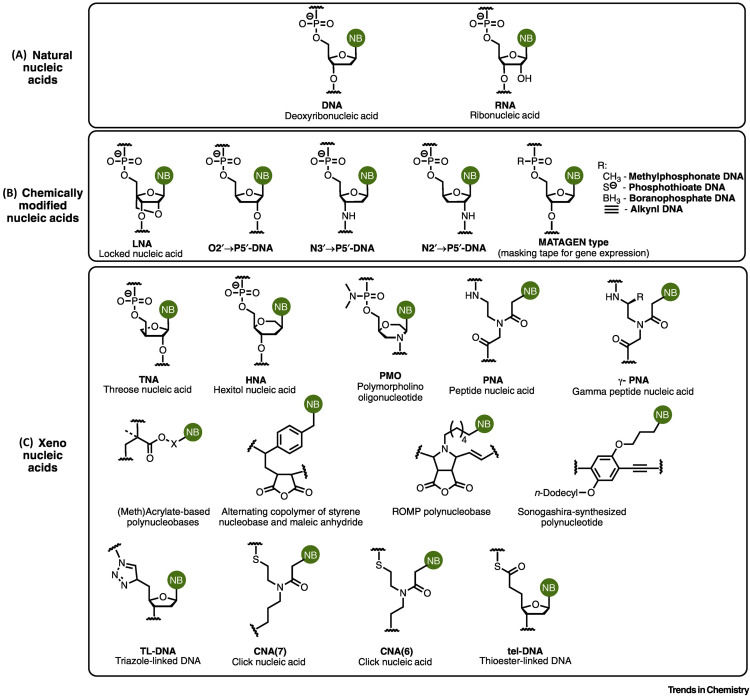
Image cited from the paper by Fairbanks et al. (7)



Comments